Let me explain you what my project is all about!
About NRNB
National Resource for Network Biology (NRNB) is an organisation which aims to advance the new science of Biological Networks through analytic tools, visualizations, databases and computing resources. In an easier words NRNB builds the technology required to assemble and analyze all kinds of protein and drug interaction, gene, cell-cell communications and similar sci-fi things you can think of. 😸
So, what is my project about ??
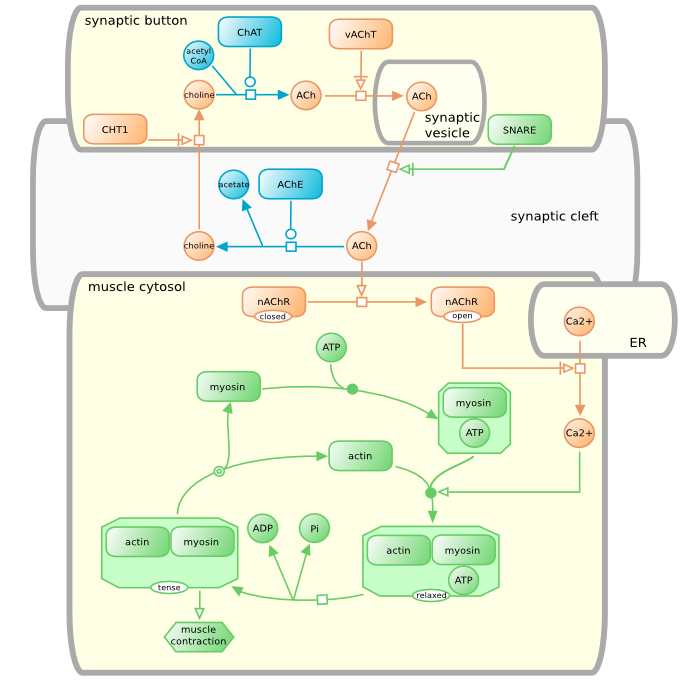
See the image above. That’s structure is built on an webapp called Newt. So what Newt basically does it helps you build, visualize and edit such patway models. All you have to do is drag and drop those boxes, connect them using arrow and name them. It’s as easy as that 😌. Cool isn’t it?
So what’s bothering Newt right now is that it doesn’t have a database of its own. User can still make those beautiful maps like the one you see above but saving those maps online isn’t one of Newt speciality. The only way you could save it is by downloading it in a format that Newt understand.
So the aim of the project is to add database support to Newt which would make it a really happy webapp.
So, what all features are you going to add to Newt ?
Well there are 2 things we plan to do.
- Deposit the Pathways to an online database.
- Query the database using algorithms such as Shortest Path between entities, K-Neighborhood, Upstream/Downstream of an entity etc to get the pathways of your interest.
Want to know about the most challenging part of this project ?
Earlier I mentioned that you would deposit the Pathway to an online database. But the thing is some part of the pathway might be already present in the database so instead of just pushing everything to the database we have to make sure that everything fits like a puzzle in the database and there’s no duplicated node/entity which is quite challenging to do as there are many rules which govern whether two nodes are different or not.
Let’s get started then 🔥🔥