Getting to know the project better.
My mentor and I already had a meeting last week, and we discussed what my targets would be for this week. So my main goals this week were trying to understand the codebase better by doing some minor changes in the code and building the database model for the Biological Maps.
So there are some things that I want to clear first. Newt is open-source software with the source code present on Github. People can deploy their own instance of Newt if they want to use it. Also, in the last blog post, I talked about integrating the database in the Newt app. Using the facility of a local database is being kept optional. By that, I mean is, the person deploying an instance of Newt will have to set some variables such as database URI to use the database (which he has deployed for storing the maps he wants to analyze).
What changes did I exactly do then?
I made some UI changes. If someone provides the database URI, some additional options to query/deposit from/to the database will appear in the toolbar/context menus. Some examples are given below.
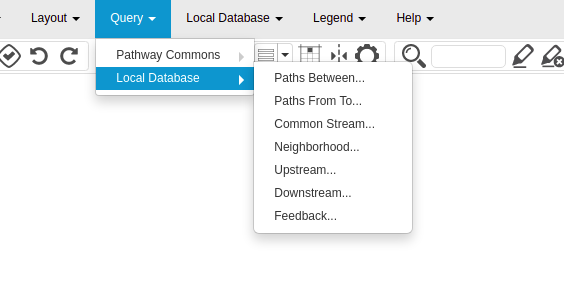
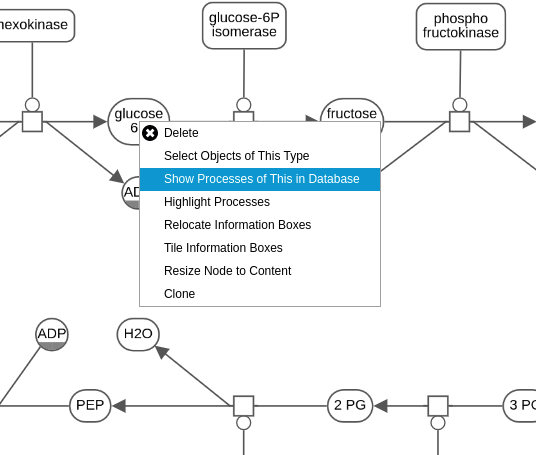
Secondly, we modeled the database for the languages PD (Process Description) and AF ( Activity Flow), which are used to build maps on Newt. We would be using the Neo4j graph database for this project. In a graph database, the entities are stored as nodes and edges. So what we did while modeling was, decide whether to store the entity as a node or an edge. Furthermore, we also modeled the properties to store in the nodes and edges, which will help us in querying and integrating the maps.